REVIEW ARTICLE
Data Analysis and Mapping of Research Interest in Clinical Trials of Tuberculosis by Text Mining Platform of Artificial Intelligence using Open-source Tool Orange Canvas
Swayamprakash Patel1, *, Ashish Patel1, Umang Shah1, Mehul Patel1, Nilay Solanki1, Mruduka Patel2, Suchita Patel3
Article Information
Identifiers and Pagination:
Year: 2022Volume: 1
Issue: 1
E-location ID: e130122200200
Publisher ID: e130122200200
DOI: 10.2174/2666782701666220113102751
Article History:
Received Date: 23/09/2021Revision Received Date: 23/09/2021
Acceptance Date: 20/10/2021
Electronic publication date: 17/03/2022

Abstract
Background: Reading every clinical trial for any disease is tedious, as is determining the current progress, especially when the number of clinical trials is huge. The Text Mining Platform of Artificial Intelligence (AI) can help to simplify the task.
Methods: A large pool of tuberculosis clinical trials has been searched through the International Clinical Trial Registry Platform (ICTRP) and used as a textual dataset. The exported dataset of 1635 clinical studies, in a comma-separated format, is preprocessed for data analysis and text mining. Data preparation, corpus generation, text preprocessing, and finally, cluster analysis were carried out using the text-mining widget of the open-source machine learning tool. The hierarchical cluster analysis was used for mapping research interests in tuberculosis clinical trials.
Conclusion: The data mining of the exported dataset of tuberculosis clinical trials uncovered interesting facts in terms of numbers. Text mining presented a total of 41 hierarchical clusters that were further mapped in twenty-five (25) different research interests among tuberculosis clinical trials. A novel technique for the rapid and practical review of major clinical trials is demonstrated. As an open-source and GUI-based tool is used for work, any researcher with working knowledge of text mining may also use this technique for other clinical trials.
1. INTRODUCTION
Tuberculosis is an ancient illness that claims millions of lives each year [1, 2]. Despite massive attempts to find the panacea, the numbers worsen. Tuberculosis with multidrug resistance (MDR) is now another problem for scientists [3-6]. Several clinical trials are registered around the globe, as well as myriads of research publications. The areas that have been explored so far need to be identified. Without this analysis, the study work cannot be directed in the right direction. This dataset is too big and continuously rising as well. Consequently, the present state of work in tuberculosis is difficult to grasp through the conventional way of literature review.
To increase the transparency in clinical trials and avoid biased or selective reporting of the results, many countries have adopted mandatory clinical trial registration prior to its execution. Furthermore, several journals have redefined their policies about the nonacceptance of research papers without the registration number for clinical trials. The WHO has also developed a shared database that gathers, compiles, and publishes meta-details of these registrations from various worldwide registries. Currently, the WHO's International Clinical Trial Registry Platform (ICTRP) has networked with 17 different country registries.
ICTRP enables registered clinical trials from many countries to be searched via the same web portal. This search platform does, however, have search and export facilities with limited filter options. Thus, one can export the entire list of the registered tuberculosis clinical trials, but it is difficult to understand the research trend between these lists without proper analysis. The ICTRP generated more than 1,600 results for a simple one-word query like “tuberculosis” (Portal was accessed in December 2019). This exported result includes fundamental pieces of clinical trial information such as Trial ID; Public Title, Scientific Title, Registration Date, Type of clinical trial, etc.
The initial statistical analysis of such data is simple. However, the whole picture of clinical trials from the exported list of clinical trials needs to be understood for a strategic review. The abundance of clinical trials makes that task complex and time-consuming. Extracting scientific information from this massive pool of clinical trials by manually reading each one of them is cumbersome. This task can be simplified by stratification or clustering these clinical trials by their common research subjects or areas. Each such cluster represents a common area of research that is explored by that particular cluster's clinical trials.
Exported ICTRP results are mostly in text form and can be considered unstructured data. The text mining technique can help in analyzing and categorizing textual data from clinical trials [7-10]. Text mining is a technique employing natural language processing (NLP) to analyze texts from both supervised (structured) and unsupervised (unstructured) datasets. For example, those textual data are titles and abstracts of biomedical literature. The text mining technique transforms these texts into meaningful numbers that can be used to analyze and understand these enormous datasets further. Text mining can help categorize or cluster these clinical trials by their common area of research.
Methods for the text mining of biomedical literature [11-14] and patents [9, 15] are explored widely. However, not much is explored using the text mining technique for clinical trials. There are very few research works that have used either complicated processes or commercial software for clinical trial text mining [16]. The majority of end-users who use such clinical trial data for their research and health-related decisions are nonprogrammers. For the text mining of clinical trials, the graphical user interface (GUI)-based and the open-source tools need to be explored.
Orange (https://orange.biolab.si/) is an open-source tool for machine learning and data visualization [17-19]. This open-source tool is available for various operating systems like Windows, Linux, and Mac. A text mining widget of the Orange tool can be utilized for text mining and further analysis of exported results from ICTRP [20].
However, similar text mining and analysis can be performed for the results obtained from other databases. For example, the clinical trial registry of the USA (www.clinicaltrials.gov) can be utilized to search clinical trials of TB in the USA. Moreover, text mining can also be performed on it. Clinical Trial Registry of India (CTRI) is also rich in terms of data. It can be accessed from the website of CTRI (www.ctri.nic.in). However, they are not providing an export facility. Manual preparation of data in Microsoft Excel® is possible in the case of CTRI.
As shown in Fig. (1), text mining and analysis involve a few necessary steps, which include data retrieval, data preprocessing, gathering of textual data (corpus), preprocessing of text, and text mining at last.
In the present work, the text mining technique is employed using an open-source and GUI-based tool – Orange. Clinical trials on TB which are retrieved from ICTRP are utilized as unstructured textual data. Research interests and trends among registered TB clinical trials are mapped using the technique of text mining. Although the present work represents the data up to 2019, one can easily export the updated dataset and can perform text mining just by replacing older data. This method is reproducible and can be used in any other dataset of clinical trials. Besides the mapping of research interest, some interesting statistics and data mining of the retrieved clinical trials are also discussed in brief.
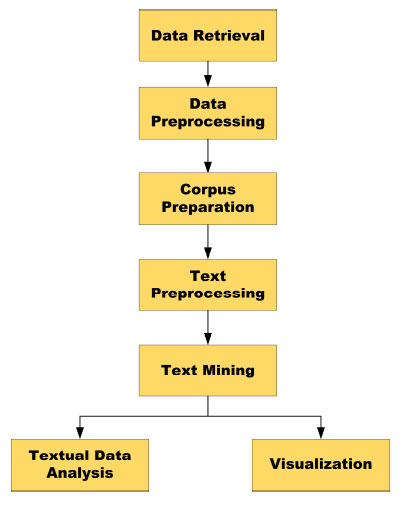 |
Fig. (1). Basic steps of text mining and analysis of textual data. |
Compendiously is a novel approach for effortlessly reviewing the massive number of literature, which is explained lucidly in this article. Text mining is a known approach among data scientists. Nevertheless, it is almost impossible to apply for nonprogrammers. A simple and GUI-based method using the open-source tool is described here using the massive data of tuberculosis clinical trials as literature.
This article is ramified into three parts. The first part is a methodology, wherein a step-by-step process is explained, covering data collection, curation, text mining, and analysis. The second part is the result section. It depicts the outcome of text mining and analysis. It basically allows readers to understand how text mining revealed the numerous information from a large pool of tuberculosis’s clinical trials. Lastly, the third part is the conclusion. It summarises information gathered from literature about tuberculosis. It also concludes the effectiveness and the easiness of the text-mining approach.
2. METHODOLOGY
2.1. Data Mining
Data Mining was performed over structured data of exported ICTRP results to understand the scenario of tuberculosis clinical trials across the globe. Various meta-analyses like country-wise and year-wise registration of clinical trials, distribution of gender among the registered clinical trials, availability of results, etc., were performed.
2.2. Text Mining
The method consists of five main steps, as shown in Fig. (1): (1) Data Retrieval; (2) Data Pre-Processing; (3) Corpus Preparation; (4) Text Pre-Processing; and finally (5) Text Mining. Microsoft Excel 2019, Orange version 3.24, Tableau 2019 , Notepad, etc., are different tools that are used for different purposes. A computer system with the Intel Core i5-7200 CPU with 2.5 GHz speed and 64-bit Windows 10 operating system was used for the entire work. The system is equipped with 8 GB of RAM to ensure smooth system function.
2.2.1. Data Retrieval and Data Pre-processing
The IRCTP portal frequently updates its database. However, IRCTP's search portal was accessed in late December 2019. Therefore, the dataset will only cover clinical trials until 2019. A single keyword query was used with the word “Tuberculosis” to search the database. All result outputs were exported in the file format of comma-separated value (.csv) and renamed “core_data.csv.” As shown in Table 1, the forty fields were exported in the result. An example (regardless of any particular clinical trial) of each field is also shown in the table. This dataset covers a total of 1635 clinical trials of tuberculosis.
Necessary data refining was done manually, without any disruption to its scientific significance. For example, “male” and “males” both are manually corrected as “Male” in the field of gender. In the same way, “Intervention” and “Interventional” both are corrected as “Interventional” in the area of the study type. Microsoft Excel was utilized for this data preprocessing, and Tableau® Desktop (Trial Version) was utilized for data analysis and visualization.
2.2.2. Corpus Preparation
Each row of “core_data.csv” represents one individual clinical trial, as shown in Fig. (2). The text mining widget's “Import Document” tool in the Orange software can handle data only in text (.txt) file format. Therefore, an individual row (from core_data.csv) must be exported to the individual text file for the preparation of the corpus. This text file must be renamed with the respective clinical trial “Trial ID.” The first row of the dataset, for example, is exported as a text file renamed as “ACTRN126100643077.txt.”
For the text mining purpose, only “Public title” and “Scientific title” were exported into the text file, which embraces the scientific meaning of the clinical trial. Thus, each text file contains the titles of its respective clinical trial. A macro function of Microsoft Excel was used for the export of each row containing only public and scientific titles in a single text file and further renaming it with its “Trial ID.” This execution has generated a total of 1635 text files, each containing the public and scientific title of their respective clinical trials.
As shown in Fig. (3), all text files, according to their source registry, were grouped into separate subfolders. These subfolders have been renamed according to their respective source registries. These sub-folders were renamed as per their respective source registry. The source registry ultimately represents an individual country where the particular clinical trial was registered. Therefore, a total of 15 subfolders with text files of their respective clinical trials were created in this way. As shown in Fig. (3), each sub-folder named after the source register represents their respective countries. All these sub-folders were stored in one principal folder named “Corpus”. This principal folder was used in the Orange software to create a corpus for further text mining.
This “Corpus” folder was browsed and selected using the Orange software's “Import Documents” tool. In Orange, each of the fifteen subfolders is referred like 15 different categories. The text files are grouped in subfolders and look like a structured dataset because they are marked with their respective “source registry.” The textual dataset in those subfolders, however, is unstructured from the perspective of text mining. The entire dataset is available from the GitHub (https://github.com/Swayamprakashpatel/Tuberculosis_Clinical_Trial_TextMining.git).
Fig. (4) shows a basic workflow for text mining in the Orange software. The Import Documents tool is connected to the Corpus tool to generate a corpus. Corpus viewer, preprocess text, bag of words, distance, hierarchical clustering, distribution, and word cloud and data table widgets were connected in series as shown in the figure.
2.2.3. Text Pre-processing
Before the analysis, various text preprocessing was performed using the “Text Preprocessing” tool in the Orange.
Table 2 shows all of the options available in the “Text Preprocessing” tool. All words were transformed into lowercase, and if present, their accents were removed. By enabling the “parse Html” option, the HTML markups were removed from its text. URLs of the text data were simply disregarded. For the tokenisation, only the word and punctuation were taken into account. Words were lemmatized using “WordNet” as a lexical database instead of mere stemming. N-gram was set to a maximum of two (N=2) words.
Besides basic stop words and regular expressions in English, some other words had also been filtered from the text. These filter words are 'tuberculosis;' 'tb;' 'exclusion;' 'inclusion;' 'criteria;' 'treatment;' 'study;' 'patient;' 'therapy;' 'medicine;' 'effect;' 'clinical;' 'trial.' These are the most common words in any clinical trial and do not constitute the scientific sense or meaning of any clinical study (at least in this work). We recommend reading the tutorials provided by the Orange software over its website for a better understanding of this tool.
2.2.4. Text Mining
For text mining, a technique of cluster analysis was used to group the clinical trials with the common research interest. The hierarchical clustering (HC) technique was utilized to understand the various research interests among the registered clinical trials of tuberculosis. Hierarchical Cluster Analysis (HCA) uses an algorithm that keeps similar objects in common groups called clusters [21-24]. Depending upon the most occurring words, they are grouped in a common cluster. The top-down strategy has been used in this work to ensure effective clustering [25]. “Distance” and “Hierarchical clustering” are the widgets in the orange software which were used for the cluster analysis. Distance between the rows and a cosine distance matrix was utilized in the hierarchical clustering. Ward's method was chosen to cluster the clinical trials [26] effectively, and the dendrogram height ratio was set at 35 percent of the total height for the selection of clusters.
| No. | Field Title | Example |
|---|---|---|
| 1. | Trial ID | ACTRN12610000643077 |
| 2. | Last Refreshed on | 15-Apr-13 |
| 3. | Public title | Effect of silymarin in the treatment of adverse effects of anti-tuberculosis drugs. |
| 4. | Scientific title | Evaluation of Silymarin in the treatment of anti-tuberculosis drug-induced hepatitis in patients newly diagnosed with tuberculosis. |
| 5. | Acronym | ALTAC |
| 6. | Primary sponsor | Government funding body National Institute of Tuberculosis and Lung Disease (NRITLD) |
| 7. | Date registration | 09/08/2010 |
| 8. | Date registration3 | 20100809 |
| 9. | Export date | 12/26/2019 9:50:07 AM |
| 10. | Source Register | ANZCTR |
| 11. | Web address | http://www.anzctr.org.au/ACTRN12610000643077.aspx |
| 12. | Recruitment Status | Recruiting |
| 13. | Other records | No |
| 14. | Inclusion age min | 18 Years |
| 15. | Inclusion age max | 100 Years |
| 16. | Inclusion of gender | Both males and females |
| 17. | Date enrolment | 01/10/2010 |
| 18. | Target size | 60 |
| 19. | Study type | Interventional |
| 20. | Study design | “Controlled: yes; Randomised: yes; Open: yes; Single-blind: no; Double-blind: no; Parallel group: yes; Cross over: no; Other: no; If controlled, specify comparator, Other Medicinal Product: yes; Placebo: no; Other: yes; Other specify the comparator: Rifampin, Pyrazinamide, Isoniazid” |
| 21. | Phase | Phase 3 |
| 22. | Countries | Outside; Iran, the Islamic Republic Of |
| 23. | Contact first name | Andre |
| 24. | Contact last name | Dr. Majid Marjani |
| 25. | Contact address | National Institute of Tuberculosis and Lung Disease, Masih Daneshvari Hospital, Dar Abad street, Niavaran, Tehran, post code:1955841452, Iran, Islamic Republic Of |
| 26. | Contact email | marjani216@hotmail.com |
| 27. | Contact tel | 9.82126E+11 |
| 28. | Contact affiliation | Aarhus University Hospital |
| 29. | Inclusion criteria | Inclusion criteria: New cases of tuberculosis |
| 30. | Exclusion criteria | Exclusion criteria: 1. Infection with Human Immune deficiency virus 2. Infection with hepatitis B virus 3. Infection with hepatitis c virus 4. pregnancy 5. Breastfeeding |
| 31. | Condition | Tuberculosis |
| 32. | Intervention | Silymarin 420 mg per day, in three doses, it will start after diagnosis of drug-induced hepatitis and stop after normalization of liver function tests, as oral tablets |
| 33. | Primary outcome | Normalization of liver function tests |
| 34. | Results date posted | 04/04/201 |
| 35. | Results date completed | 31/10/2019 |
| 36. | Results URL link | https://clinicaltrials.gov/ct2/show/results/NCT00814671 |
| 37. | Retrospective flag | Yes |
| 38. | Bridging flag true false | FALSE |
| 39. | Bridged type | Parent |
| 40. | Results yes no | yes |
 |
Fig. (2). Core data in comma-separated (.csv) file format (only the first few columns and rows shown here). |
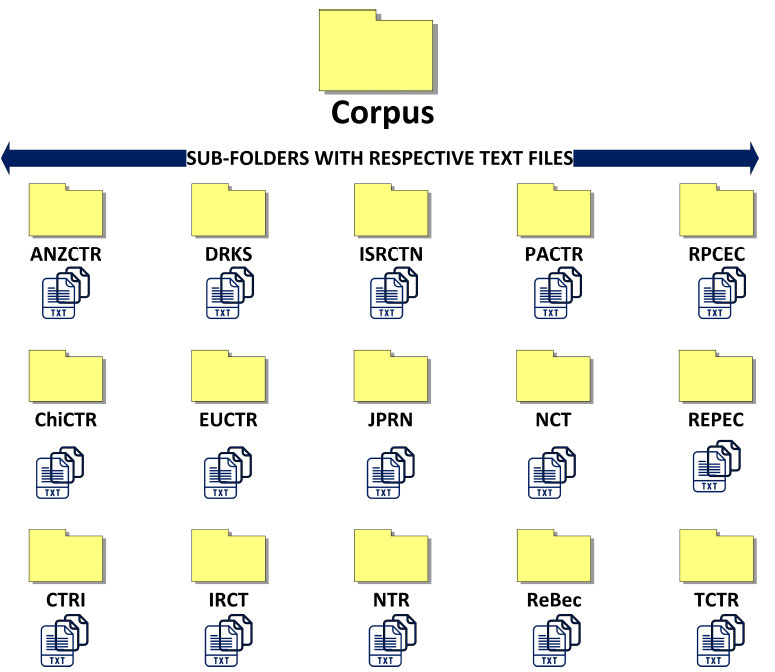 |
Fig. (3). Grouping of text files of the clinical trial as per respective source register in appropriate folders. |
 |
Fig. (4). Exemplary workflow of text mining in orange. |
| Pre-processing Feature | Options | Status |
|---|---|---|
| Transformation | Lowercase | Enabled |
| Remove Accents | Enabled | |
| Parse HTML | Enabled | |
| Remove URLs | Enabled | |
| Tokenization | Word and Punctuation | Enabled |
| Whitespace, Sentence, Regexp, Tweet | Disabled | |
| Normalization | WordNet Lemmatizer | Enabled |
| Porter stemmer, Snowball stemmer, UDPipe Lemmatizer | Disabled | |
| Filtering | Stop word (English) | Enabled |
| Stop word (Tuberculosis Word list.txt) | Enabled | |
| Regexp | Enabled | |
| Lexicon, Document Frequency, Most Frequent Token | Disabled | |
| N-gram range | 1-2 | Enabled |
As shown in Error! Reference source not found., the “hierarchical cluster” (HC) tool was connected to the “Distribution,” “Word Cloud,” “Data Table,” and “Contingency Table” tools in the series manner. “Selected Data-to-Data” and “Selected Data-to-Corpus” options were set as the connection between HC, distribution, and word cloud tools, respectively. At the same time, “corpus-to-data” and “data-to-data” connections were utilized for word cloud, data table, and contingency table tools. For the analysis of each cluster, the most common words were selected from the word cloud one by one until it covered all clinical trials in the contingency table. This list of words represents that particular cluster (Refer to Supplementary Table 1). The meaningful research area was concluded manually from this list of words and considered as a common research interest of the clinical trials within the respective cluster.
In some clusters, the resolution of words for firm mapping of research interest is poor. We can not conclude a common research interest from their list of most frequent words. In such cases, it is imperative to perform sub-clustering. The process of sub-clustering is simple and similar to hierarchical clustering. It includes the selection of clusters from the distribution tool for which sub-clustering is required. Connect the distribution widget further to the new hierarchical clustering widget, followed by a new distribution widget and a new word cloud widget in series. Mapping of research interest can be further performed for the subclusters in a way similar to that performed for its parent cluster.
3. RESULTS AND DISCUSSION
3.1. Data Mining
As shown in Fig. (5), the United States has made significant contributions, among others, in terms of the number of clinical trials on tuberculosis. The Year-wise contribution (Fig. 6) of clinical trials on tuberculosis also revealed a steady increase in the number of registrations in India and China. In the US, the number of registrations is more compared to other countries, but this rise in the registration of clinical trials has stagnated over the last couple of years.
The majority of the clinical trials are not specific to gender (Table 3). Trials included both male and female volunteers. However, data reveals that few of the clinical trials are gender-specific, and the trial includes volunteers of just a specific gender. Data were exported separately for “only male” and “only female” and processed for text mining. However, as this number is minimal, they can be studied individually. Of India's 127 clinical trials, none showed any gender-specific study. It may be because gender is not specified in India's clinical trials, or there is an issue at the ICTRP level in data retrieval. The possibility of the latter is high.
Analysis of the availability of results as “yes” or “no” in the data revealed the shocking fact of clinical trials on tuberculosis (Fig. 7). Only 87 of the registered clinical trials are available with their results, out of 1635 registered clinical trials. It may also conclude that only 5.32 percent of registered clinical trials are completed. Even if we disregard the newly registered clinical trials of the last five years (2015 to 2019), only 73 (8.1 percent) of the 901 registrations by 2014 have been completed, and their results are available at IRCTP. A news article [27] from the American Association for the Advancement of Sciences (AAAS) posted by ScienceMag ® also criticizes similar findings. The unavailability of clinical trials’ results was also questioned at different platforms [28, 29]. This conclusion, however, is based solely on data exported from IRCTP. At the ICTRP level, there could be a significant chance of error in the data. We found a few ChiCTR clinical trials, the results of which are available on their portal but are not reflected or updated as “yes” in ICTRP. Such an error needs to be addressed and resolved imperatively. Otherwise, the ICTRP's crux motive will eventually be compromised. Even if such an error may exist at the end of ICTRP, it is still true that the percentage of completed clinical trials is substantially low compared to total registered clinical trials. In our independent search at clinicaltrials.gov (US National Library of Medicine), we found that by the end of 2019, only 98 trials had been tagged as completed out of 1055 registered clinical trials.
In clinical trials, the majority of types of studies are either interventional or observational. However, the types of studies are not limited to just these two. Chinese clinical trials represent a gamut of study types other than these two. These include diagnostic testing, epidemiological research, research into relative factors, basic science, research on health services, the study of treatment, screening, research on prognosis, prevention, and causes. To understand the exact meaning of these types of studies, one must explore the Chinese Clinical Study Registry website. In each country, the number of interventional studies is much more than the number of observational or other studies, as shown in Fig. (8)
Entries in the column labeled as “phase” were refined manually using Microsoft Excel®. This manual refinement was required as multiple aliases were utilized for the same phase of the clinical trial. For instance, “Phase-4”; “Phase4”; “Phase IV”; “IV” and simply “4” were utilized to represent phase - 4. Many other terms, like basic science, other, phase 0, pilot study, and post-market, are also found in this column, commonly regarded as “Other” in Table 4. This analysis reveals that only 7.46 percent of total registered clinical trials are phase-4 trials. Particularly for India, the percentage of phase-4 trials is higher than in other phases. It seems optimistic and hopeful for a meaningful outcome. However, none of these eight clinical trials has reported any results yet. The Trial IDs of them are: (1) CTRI/2009/091/000476; (2) CTRI/2009/091/000511; (3) CTRI/2012/06/002742; (4) CTRI/2012/11/003088; (5) CTRI/2012/11/003155; (6) CTRI/2013/07/003830; (7) CTRI/2017/04/008329; (8) CTRI/2017/09/009693.
 |
Fig. (5). Country (source register) wise contribution to tuberculosis clinical trials. |
 |
Fig. (6). Year-wise contribution of each country to tuberculosis clinical trials. |
| Source Register |
Number of Registrations n (% of Total) |
Inclusion Gender | |||
|---|---|---|---|---|---|
|
Only Male |
Only Female |
Both |
Not Specified |
||
| ANZCTR | 26 (1.59) | 1 | 1 | 24 | - |
| ChiCTR | 132 (8.07) | 6 | 1 | 122 | 3 |
| NCT | 1008 (61.65) | 28 | 30 | 950 | - |
| CRIS | 15 (0.92) | - | - | 15 | - |
| CTRI | 127 (7.77) | - | - | - | 127 |
| EUCTR | 39 (2.39) | - | - | 39 | - |
| DRKS | 6 (0.37) | - | - | 6 | - |
| IRCT | 24 (1.47) | 1 | - | 23 | - |
| ISRCTN | 103 (6.03) | - | 2 | 97 | 4 |
| JPRN | 39 (2.39) | - | - | 39 | - |
| NTR | 4 (0.24) | - | - | - | 4 |
| PACTR | 59 (3.61) | 2 | 1 | 56 | - |
| REBEC | 10 (0.61) | 1 | - | - | 9 |
| REPEC | 26 (1.59) | - | - | 15 | 11 |
| TCTR | 17 (1.04) | - | - | 17 | - |
 |
Fig. (7). Country-wise availability of results for registered clinical trials on tuberculosis. |
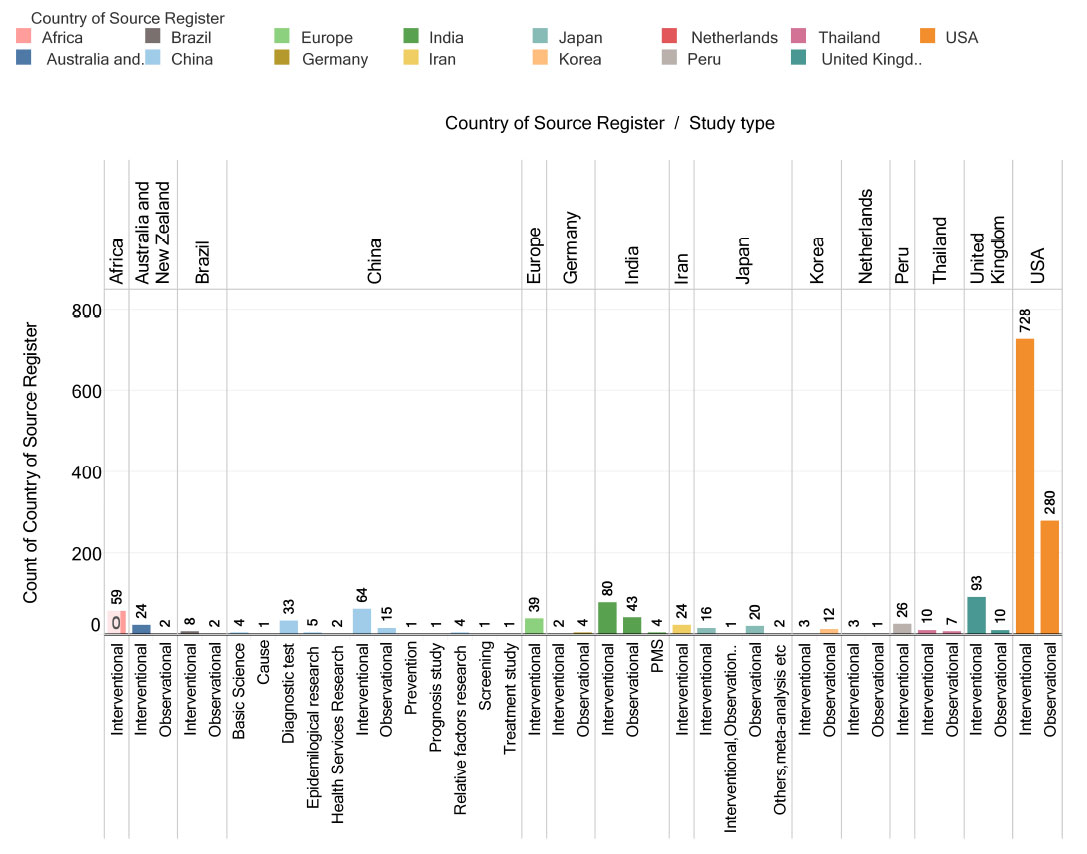 |
Fig. (8). Country-wise study of tuberculosis clinical trials. |
| Country of Source Register |
% Phase of total clinical trial (% Phase of the total clinical trial of the individual country) |
||||
|---|---|---|---|---|---|
| Phase-4 | Phase-3 | Phase-2 | Phase-1 | Other | |
| Africa | 0.06 (1.69) | 0.12 (3.38) | 0.36 (10.1) | 0.06 (1.69) | 2.99 (83.0) |
| Australia and New Zealand | 0.18 (11.5) | 0.18 (11.5) | 0.06 (3.84) | 0.00 (0.00) | 1.16 (73.0) |
| Brazil | 0.12 (20.0) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.48 (80.0) |
| China | 0.48 (6.06) | 0.12 (1.51) | 0.06 (0.75) | 0.55 (6.81) | 6.85 (84.8) |
| Germany | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.06 (16.6) | 0.30 (83.3) |
| Europe | 0.00 (0.00) | 0.30 (13.5) | 0.48 (21.6) | 0.00 (0.00) | 1.46 (64.8) |
| India | 0.48 (6.20) | 1.10 (13.9) | 1.03 (13.1) | 0.42 (5.42) | 4.83 (61.2) |
| Iran | 0.06 (4.16) | 0.30 (20.8) | 0.00 (0.00) | 0.00 (0.00) | 1.10 (75.0) |
| Japan | 0.12 (5.12) | 0.06 (2.56) | 0.06 (2.56) | 0.06 (2.56) | 2.07 (87.1) |
| Korea | 0.06 (6.66) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.85 (93.3) |
| Netherland | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.00 (0.00) | 0.24 (100) |
| Peru | 0.30 (19.2) | 0.67 (42.3) | 0.61 (38.4) | 0.00 (0.00) | 0.00 (0.00) |
| Thailand | 0.12 (11.7) | 0.30 (29.4) | 0.12 (11.7) | 0.00 (0.00) | 0.48 (47.0) |
| United Kingdom | 0.18 (2.91) | 0.42 (6.79) | 0.06 (0.97) | 0.06 (0.97) | 5.56 (88.3) |
| USA | 5.25 (8.53) | 6.72 (10.9) | 12.1 (19.7) | 11.7 (19.0) | 25.7 (41.7) |
| Total % of Phase | 7.46 | 10.3 | 15.0 | 12.9 | 54.1 |
3.2. Text Mining
As shown in Fig. (9), a huge pool of 17,825 words with 31,920 tokens from the corpus of clinical trials for tuberculosis were generated after preprocessing of text. In this pool, the word “HIV” (n=519) has the highest count, followed by the words “pulmonary” and “phase” (n=510 and 484), respectively. Similarly, it was found that a total of 173 drugs are listed in clinical trials after appropriate preprocessing for the drugs involved in the clinical trials (Fig. 10). The most common name for the drug is “isoniazid,” followed by “rifampicin” and “rifapentine.”
3.2.1. Hierarchical Cluster Analysis
As shown in Fig. (11), a total of 41 clusters were identified and selected for further analysis via the hierarchical clustering technique.
The supplementary table (Table 1) in the appendix represents the research interest of clinical trials of every cluster in a comprehensive manner. Out of 41 clusters, only seven clusters need sub-clustering to understand the research interest of their clinical trials. Thirty-three (33) clusters, for which successful mapping of the research area was performed, covering 1015 (Supplementary Table 2) clinical trials out of a total of 1635. The remaining 620 clinical trials, consisting of C27, C29, C32, C34, C35, C37, and C40, may be mapped by sub-clustering for their research interest. This sub-clustering and analysis are not shown here since it can extend the usual publication limit.
In the supplementary table (Table 1), the research interests or areas mapped for 1015 clinical trials are summarized and shown in Table 5. As per this summary, the highest number of TB clinical trials are focused on HIV infection. In addition to common research trends on HIV, vaccine, BCG, MDR, etc., other interesting research trends including vitamins, active case findings in the community, diabetes, pleural effusion, fixed-dose combinations, meningitis, etc. are also mapped.
Cluster 35 contains 384 clinical trials for which the mapping of research interest is not possible without sub-clustering. Sub-clustering of C35 (similar to hierarchical clustering) and the selection of 80% of the total height of the dendrogram have grouped these 384 clinical trials in 14 different Sub-clusters. As shown in Fig. (12), these 14 subclusters have a positive silhouette score. It means the mapping of research areas for these clusters is possible with excellent resolution. The research area of 8 clusters out of these 14 was identified and listed in Table 6. Rest can be easily mapped through re-reclustering (not shown here).
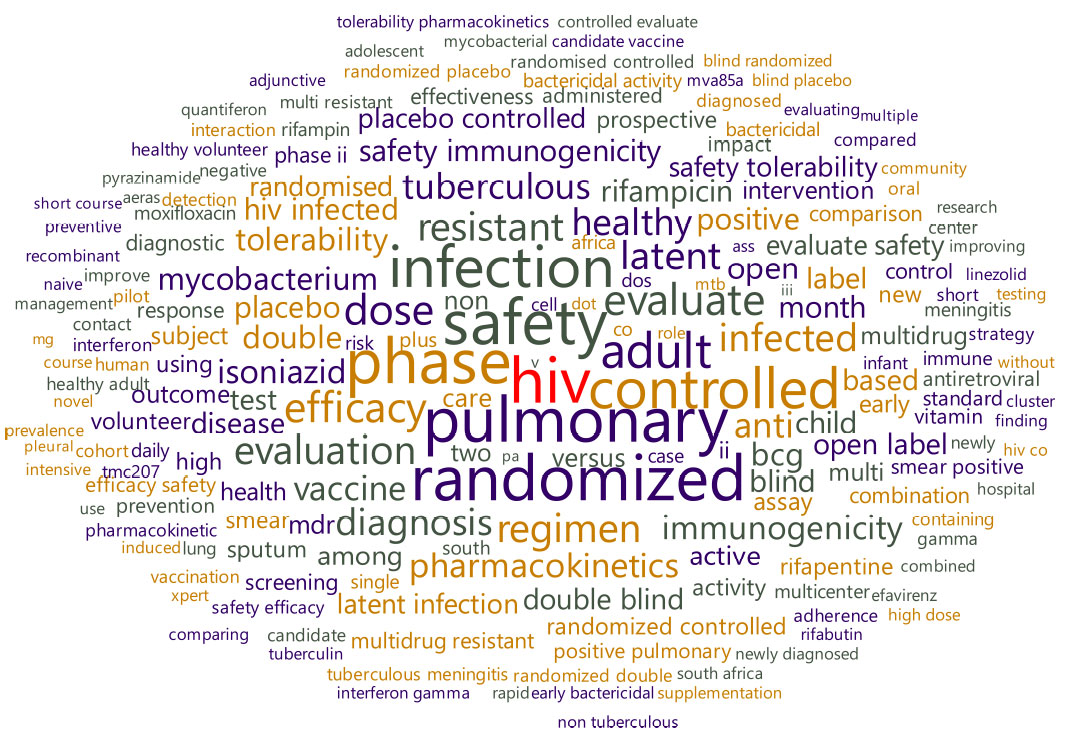 |
Fig. (9). Word cloud of text from tuberculosis clinical trials. |
 |
Fig. (10). Word cloud of drugs mentioned in tuberculosis clinical trials. |
 |
Fig. (11). Hierarchical cluster distribution. |
| Sr. No | Common Research-Interest / Trend / Area, Which Represents the Entire Cluster | Number of Clinical Trials* |
|---|---|---|
| 1. | Vaccine and Immunological Studies | 234 |
| 2. | Placebo related studies | 56 |
| 3. | BCG related studies | 20 |
| 4. | HIV and other co-infection related studies | 292 |
| 5. | MDR related studies | 104 |
| 6. | Bacteriocidal activities of compound | 31 |
| 7. | Regimen related studies | 42 |
| 8. | Pulmonary tuberculosis related studies | 42 |
| 9. | Diabetes and tuberculosis related studies | 12 |
| 10. | Diagnostic related studies | 86 |
| 11. | Latent tuberculosis related studies | 45 |
| 12. | Meningitis related studies | 27 |
| 13. | Non-tuberculosis Mycobacteria related studies | 24 |
| 14. | Plural effusion related studies | 19 |
| 15. | Recombinant products related studies | 12 |
| 16. | Isoniazid related studies only | 28 |
| 17. | Rifapentine, Moxifloxacin, and Rifabutin related studies | 20 |
| 18. | Pharmacokinetic and toxicity of various anti-TB agents | 104 |
| 19. | Directly Observed Therapy (DOT) related studies | 22 |
| 20. | Fixed-Dose Combination related studies | 21 |
| 21. | Role of Vitamins related studies | 20 |
| 22. | Studies related to Active Case Finding in various communities | 13 |
| 23. | Quality of Health and Life-related studies | 48 |
| 24. | Xpert MTB-RIF assay related studies | 19 |
| 25. | lipoarabinomannan (LAM) assay related studies | 06 |
(* numbers of clinical trials in a particular cluster).
 |
Fig. (12). Silhouette plot of subclusters of C35. |
| Sr. No | Common Research-Interest / Trend / Area, Which Represents the Entire Cluster | Number of Clinical Trials* |
|---|---|---|
| 1. | Early mortality related studies | 4 |
| 2. | Low birth weight / weight (in TB) related studies | 8 |
| 3. | Diagnostics related studies | 14 |
| 4. | Screening and tracing related studies | 26 |
| 5. | Smoking-related studies | 6 |
| 6. | South-africa specific studies | 11 |
| 7. | Cohort Studies | 8 |
| 8. | Surgeries and Tuberculosis related studies | 8 |
CONCLUSION
Data Mining
Overall, data mining suggests that clinical trials for tuberculosis should be focused more on the new drug, new regimen, and more specifically on MDR-TB to be prepared for the possibly upcoming disaster with TB. It is also crucial to publish the results of every registered clinical trial as soon as possible to boost knowledge about tuberculosis in the research community. A provision should be explored regarding the current status of the registered clinical trial and the probable date of its completion. Countries other than the US must considerably increase their contribution to tuberculosis research and clinical trials.
Text Mining
Comprehension of the research pattern manually from a large number of clinical trials is a complicated job. Text mining is indeed a convenient approach for understanding any disease from registered clinical trials. For the research fraternity with nonprogramming background, the use of GUI-based open-source software for text mining and clinical trial analysis is quite simple. The research-interest mapping method explained here can be easily reproduced for any clinical trial dataset exported from ICTRP and other registries. This mapping technique can enable researchers to understand the overall areas of research where clinical trials are initiated. In this work, we have mapped 23 different research areas/topics in which tuberculosis clinical trials are registered worldwide. The conventional method of manually reviewing pieces of literature cannot be employed when the number is large. It is even tedious and intricate to review literature like clinical trials if their numbers are exceedingly more than a hundred. This article reviews over 1600 pieces of literature in order to obtain an overview. In essence, text-mining is the best approach to perform a literature review. The GUI-based open-source tool can be easily used by researchers with a non-programming background.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.
APPENDIX
VBA CODE FOR EXPORT OF EACH ROW IN .TXT FILE FORMAT
Sub SaveRowsAsTXT()
Dim wb As Excel.Workbook, wbNew As Excel.Workbook
Dim wsSource As Excel.Worksheet, wsTemp As Excel.Worksheet
Dim r As Long, c As Long
Dim filePath As String
Dim fileName As String
Dim rowRange As Range
Dim cell As Range
filePath = “C:\Tuberculosis\”
For Each cell In Range(“B1”, Range(“B10”).End(xlUp))
Set rowRange = Range(cell.Address, Range(cell.Address).End(xlToRight))
Set wsSource = ThisWorkbook.Worksheets(“Sheet1”)
Application.DisplayAlerts = False 'will overwrite existing files without asking
r = 1
Do Until Len(Trim(wsSource.Cells(r, 1).Value)) = 0
ThisWorkbook.Worksheets.Add ThisWorkbook.Worksheets(1)
Set wsTemp = ThisWorkbook.Worksheets(1)
For c = 2 To 25
wsTemp.Cells((c - 1) * 2 - 1, 1).Value = wsSource.Cells(r, c).Value
Next c
fileName = filePath & wsSource.Cells(r, 1).Value
wsTemp.Move
Set wbNew = ActiveWorkbook
Set wsTemp = wbNew.Worksheets(1)
wbNew.SaveAs fileName & “.txt”, xlTextWindows 'save as .txt
wbNew.Close
ThisWorkbook.Activate
r = r + 1
Loop
Application.DisplayAlerts = True
Next
End Sub
REFERENCES
| [1] | Barberis I, Bragazzi NL, Galluzzo L, Martini M. The history of tuberculosis: from the first historical records to the isolation of Koch’s bacillus. J Prev Med Hyg 2017; 58(1): E9-E12. |
| [2] | Global Tuberculosis Report; World Health Organization: Geneva, 2019. Available from: https://www.who.int/publications/i/item/9789241565714 |
| [3] | Migliori GB, Tiberi S, Zumla A, et al. MDR/XDR-TB management of patients and contacts: Challenges facing the new decade. The 2020 clinical update by the global tuberculosis network. Int J Infect Dis 2020; 92S: S15-25. |
| [4] | Young M, Craig J. Urgent global action is needed on multi drug-resistant tuberculosis (MDR-TB)–can small cone moxa contribute to a global response? Eur J Integr Med 2020; 37: 101072. |
| [5] | Li BY, Shi WP, Zhou CM, et al. Rising challenge of multidrug-resistant tuberculosis in China: a predictive study using Markov modeling. Infect Dis Poverty 2020; 9: 65. |
| [6] | Guglielmetti L, Low M, McKenna L. Challenges in TB regimen development: preserving evidentiary standards for regulatory decisions and policymaking 2020. |
| [7] | Korhonen A, Séaghdha DO, Silins I, Sun L, Högberg J, Stenius U. Text mining for literature review and knowledge discovery in cancer risk assessment and research. PLoS One 2012; 7(4): e33427. |
| [8] | Fabbri S, Elis H, Andre DT, Anderson B, Augusto Z, Cleiton S. Using information visualization and text mining to facilitate the conduction of systematic literature reviews. International Conference on Enterprise Information Systems Springer Berlin, Heidelberg. 2012; 243-56. |
| [9] | Rodriguez-Esteban R, Bundschus M. Text mining patents for biomedical knowledge. Drug Discov Today 2016; 21(6): 997-1002. |
| [10] | Przybyła P. Text mining resources for the life sciences. Database (Oxford) 2016. |
| [11] | Zhu F, Patumcharoenpol P, Zhang C, et al. Biomedical text mining and its applications in cancer research. J Biomed Inform 2013; 46(2): 200-11. |
| [12] | Fleuren WWM, Alkema W. Application of text mining in the biomedical domain. Methods 2015; 74: 97-106. |
| [13] | Saffer JD, Burnett VL. Introduction to Biomedical Literature Text Mining: Context and Objectives.Biomedical Literature Mining 2014; 1-7. |
| [14] | Simon C, Davidsen K, Hansen C, Seymour E, Barnkob MB, Olsen LR. BioReader: a text mining tool for performing classification of biomedical literature. BMC Bioinformatics 2019; 19(Suppl. 13): 57. |
| [15] | Senger S. Assessment of the significance of patent-derived information for the early identification of compound-target interaction hypotheses. J Cheminform 2017; 9(1): 26. |
| [16] | Korkontzelos I. Text mining for efficient search and assisted creation of clinical trials. Proceedings of the ACM 5th international workshop on Data and text mining in biomedical informatics 2011; 43-50. |
| [17] | Demšar J. Orange: data mining toolbox in Python. J Mach Learn Res 2013; 14(1): 2349-53. |
| [18] | Jovic A, Brkic K, Bogunovic N. An overview of free software tools for general data mining. 2014 37th International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO) 2014; 1112-1117. |
| [19] | Kaur MP, Rana QP. Advances in Agri-Management. OrangeFuture Challenges 2017; 155. |
| [20] | Demšar J, Zupan B. Orange: Data mining fruitful and fun-a historical perspective. Informatica (Vilnius) 2013; 37(1) |
| [21] | Ghosal A. A short review on different clustering techniques and their applications.Emerging Technology in Modelling and Graphics 2020; 69-83. |
| [22] | Zou HJWPC. Clustering algorithm and its application in data mining. Wirel Pers Commun 2020; 110(1): 21-30. |
| [23] | Demšar J. Orange: data mining toolbox in Python. J Mach Learn Res 2013; 14(1): 2349-53. |
| [24] | Kubek M. Natural Language Processing and Text Mining 2020. |
| [25] | Rokach L, Maimon O. Clustering Methods.Data Mining and Knowledge Discovery Handbook 2005; 321-52. |
| [26] | El-Hamdouchi A, Willett P. Hierarchic document classification using Ward’s clustering method. Proceedings of the 9th annual international ACM SIGIR conference on Research and development in information retrieval Association for Computing Machinery New York, NY, USA. 1986; 149-56. |
| [27] | Piller C. FDA and NIH let clinical trial sponsors keep results secret and break the law 2020. Available from: https://www.sciencemag.org/news/2020/01/fda-and-nih-let-clinical-trial-sponsors-keep-results-secret-and-break-law |
| [28] | Fleming N. Top US institutes still aren’t reporting clinical-trial results on time 2019. Available from: https://www.nature.com/articles/d41586-019-00994-1 |
| [29] | Piller C. Transparency on trial. Science 2020; 367(6475): 240-3. |




